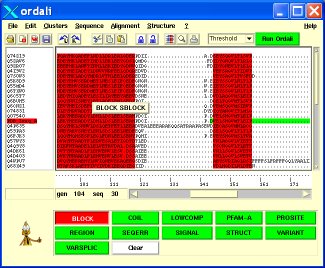
All the different menus are described in the Menus section.
The icon bar gives shorcuts to some of the menus items. This bar also contains the "Run Ordalie" button which launches residue conservation calculation using the method selected through the pull-down menu placed at its left. The methods are describged in the "Identification of conserved residues" section.
The sequences names are displayed on the left part of the frame. The sequences names highlighted in red correspoind to PDB sequences. By adding/removing separators, the user can customize its own clusters of sequences, that can subsequently be used for residues conservation identification. Sequence order can be changed using the Cut/Copy/Paste mechanism.Upon certain options, sequences names colors can be changed,i.e. when invoking the Phylum clustering. In the case of Macsims like alignments, the sequence definition will be displayed above the mouse pointer.
The right part of the frame contains the alignment itself. Moving the mouse pointer over a sequence will display both the position relative to the sequence and to the alignment at the bottom left of the alignment frame.
When alignment features are available, the alignment will be colored according to this features, and moving the mouse over the feature will display the note associated to it, for example, in the case of a PFAM domain, the describtion of the domain will be shown.