|
Written by Administrator
|
|
Tuesday, 20 May 2014 11:09 |
Translational informatics
Development of a computational infrastructure dedicated to the integrated analysis of data from "-omics" sciences (cf. www.omics-ethics.org/ fr/definition-science-omics) related to human genetic diseases and health. This infrastructure includes algorithms and original methods, such as:
- Algorithms for integrative structural biology, in the context of ongoing funded projects, including the European INSTRUCT research infrastructure for 'Integrative Structural Biology' and the French ANR Investments for the Future project: FRISBI, coordinated by Bruno Klaholtz, IGBMC, Strasbourg.
- Multi-scale methods for a better understanding of protein properties (structure, interactions, dynamics...) and better integration of the heterogenous data associated with a protein or a family of proteins. Ongoing project, BIPBIP, funded by French ANR in collaboration with Annick Dejaegere, IGBMC, Strasbourg.
- Evolutionary analyses of protein sequences from NGS data, applied to health, in collaboration with the Data_Mining data mining and SONIC teams.
- GREMSAP (GRid Evolutionary Multiple Sequence Alignment Platform), in collaboration with the SONIC team, l’Institut des Systèmes Complexes Paris Île de France, l’Institut de Neurobiologie Albert Fressard.
- Social Network Clinical Database for Intellectual Disabilities: Development of a social network for patients, related to genetic diseases causing developmental disabilities, in collaboration with the SONIC team, the Translational medicine & neurogenetics team, IGBMC, led by Jean-Louis Mandel, and the [http:/www.ucad.sn Cheikh Anta Diop University, Senegal].
- Infrastructure for 'big data' management for the translational analysis of mutations involved in human genetic disease. BIRD/SM2PH-central is aimed at the integration of heterogeneous data (genomic, phenotypic, evolution, cellular networks,...), with data mining methods (association rules, inductive logic programming,...), and includes the design of a semantic query language (BIRD-QL) and the development of original graphical interfaces. These developments are done in collaboration with Hoan Ngyugen, IGBMC, Strasbourg.
Systems bioinformatics
Development of research in the field of biological systems analysis, to understand genotype-phenotype relationships, notable concerning genetic diseases, and the study of complex biological systems, for example in various cancers or rare diseases (ciliopathies, myopathies,...) in close collaboration with the Human Genetics Laboratory, led by Hélène Dollfus (www.unistra.fr/index.php?id=19264&L=3). The originality of our approach lies in the extraction and exploitation of evolutionary information (sequence analysis, comparative genomics, etc.) in order to improve the analysis of the different levels of complexity in biological systems. Some examples of systems-level projects:
- Analysis of macromolecular complexes (TFIID...) in collaboration with the Department of Integrative structural biology, IGBMC.
- Analysis of organelles (exosome, endosome...) in collaboration with the membrane traffic and lipid signaling team, GMGM: synthetic yeast evolution, the characterization of the shared / specific features at different levels (nucleotide, gene, pathway), the identification of correlations between the evolutionary scenarios and the modifications identified in the molecular processes.
- Genomic analysis of 1000 myopathy patients (Myocapture project and ongoing FRM-funded project, in collaboration with le team of Jocelyn Laporte, IGBMC) with the goal of identifying and characterizing new genes in these diseases.
- Analysis of genomic data from ciliopathy patients (Bardet-Biedl Syndrome and Alström Syndrome) to identify the genes responsible for the diseases, and to understand the links between genotypes/phenotypes, in collaboration with Human Genetics Laboratory, led by Hélène Dollfus.
- Integration of "omics" data to develop a new generation of the Vaccinia virus (VACV). Ongoing funded project (OncoVaccine: ANR Recherches Partenariales et Innovation Biomédicale) in collaboration with TRANSGENE, the HTCS platform led by Laurent Brino, IGBMC and the team of Etienne Weiss, IREBS, Strasbourg.
- ImAnno is a collaborative project between the LBGI and teams at the IGBMC, Strasbourg (P. Dollé) and the Institute of Vision, Paris (J. Sahel, T. Leveillard) aimed at developing an ergonomic and integrative annotation tool for biological images (ISH, fundus...). Once annotated, the images allow to access all the knowledge extraction tools implemented or developed by the LBGI, including protein-protein interaction networks, tools for automatic analysis of the evolution, transcriptomic data, etc.
- Multi-scale tempo-spatial modeling of the primary cilia as a means of communication (intracellular transport, physical cell-cell interactions, environmental sensory system) and its role in the cell cycle, development, as well as the evolution of eukaryotic organisms, in collaboration with the Digital Campus for Complex Systems.
|
|
Last Updated on Tuesday, 20 May 2014 11:44 |
|
Integrative bioinformatics for the analysis of complex systems implicated in human diseases
Modern biology is characterized by huge volumes of complex, heterogeneous and noisy data resulting from i) the inherent variability of living organisms, ii) the low reproducibility of high throughput technologies and iii) the wide variety of computational and statistical analyses. This impressive flood of data means that the bioinformatics field must now evolve to isolate and filter valuable knowledge from the noise, errors or redundancy. This requires the combination of complementary automated data analysis and prediction tools with efficient storage and querying of constantly updated data. Such integrated approaches are an essential prerequisite to take full advantage of the opportunities provided by modern biology to unravel the principles and mechanisms of life and introduce them in a useful modeling of complex systems.
We focus on the development of robust, automated and integrated in silico approaches to study the evolution and behavior of complex biological systems (“hyperstructures”, networks, etc) in human diseases. Our major research axis concerns the exploitation of Evolution as it captures the physical, spatial, environmental and historical constraints at work in living systems and thus, if correctly deciphered, can provide a unique framework for filtering noise and for identifying shared or specific features, and subsequently propagating the inferred knowledge between biological systems. Such evolutionary inference approaches now play an important role in most high throughput studies: genome annotations, promoters, transcriptomes, proteomes or interactomes. Although many in silico tools exist, they remain largely underutilized due to the lack of a proper informatics infrastructure to make these approaches robust, accessible and easy to use in a high data flow context.
Our global strategy involves the creation of an iterative cycle combining computational developments (algorithms, databases…) to facilitate high quality human expert analysis of complex animal systems, which in turn are used to validate and refine the computational strategies.
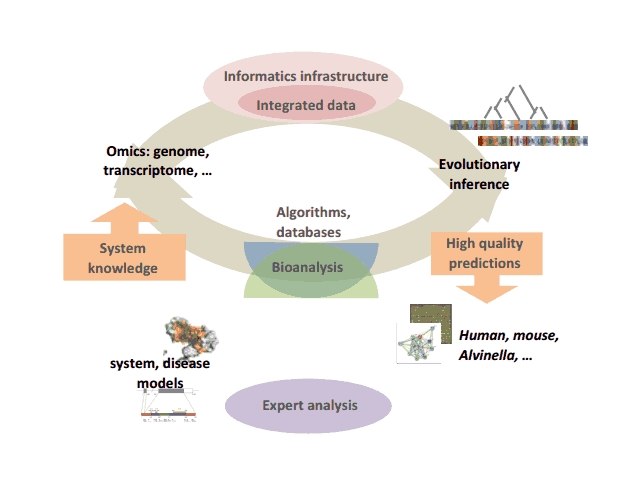
Iterative cycle of computational development and bioanalysis
At the computational level, we are addressing several problems related to this strategy:
- the design of novel algorithms, cascades of automated functional genomics data analysis, benchmark databases and expert systems for the understanding and exploitation of evolutionary information. The methodologies are developed and evaluated in the context of well dominated biological systems encompassing various levels of complexity (macromolecular structural complex, networks, diseases…) in Alvinella pompejana, mouse or human,
- the development of the BIRD (Biological Integration and Retrieval of Data) system, specifically designed to automatically generate databases managing and integrating heterogeneous local or remote biological data (genomics, transcriptomics, proteomics...). BIRD provides the core for the SM2PH-db (from Structural Mutation to Pathology Phenotypes in Human-database) aimed at studying, for all human monogenic diseases, the structural impact of genetic changes in the framework of evolutionary, functional and phenotypic information.
At the bioanalytical level, taking advantage of our integrated computational approaches and within the framework of longstanding international, national or local collaborations, we are participating in the analysis of complex systems implicated in human diseases. This involves notably, the study of functional impairment related to retinal or brain diseases, the identification of mutational patterns related to the Bardet-Biedl Syndrome and the characterization of genomic and transcriptomic context in various cancers. |
Development of bioinformatics software:
- Alvinella consortium (12 european teams)
- SPINE2 Consortium (20 european teams)
- GJ. Barton; Univeristy of Dundee, Scotland
- P. Collet; N. Lachiche; ICube, Strasbourg
- A. Dejaegere; IGBMC, Strasbourg
- MD. Devignes; LORIA, Nancy
- MH. Facci, AM. Armbuster, I. Sauvage; IBM France
- TJ. Gibson; EMBL, Heidelberg, Germany
- DG. Higgins; University College Dublin, Ireland
- Mayer; Institut Pasteur, Paris
- P. Jollivet; Station Biologique de Roscoff
- P. Koehl; UC Davis, USA
- G. Labesse; CBS, Montpellier
- C. Marino Buslje; Uni. Buenos Aires, Argentina
- P. Pontarotti; Marseille
- E. Pontelli; University of New Mexico, USA
- AK. Royyuru; IBM Watson, USA
- P. Schultz; IGBMC, Strasbourg
- A. Stoltzfus; NIST, USA
- A. Van Dorsselaer; IPHC, Strasbourg
Database development:
- P. Bork; EMBL, Heidelberg, Germany
- P. Dollé; IGBMC, Strasbourg
- W. Krezel; IGBMC, Strasbourg
- A. Pujol; Barcelona University, Spain
Ciliopathies:
- H. Dollfus; UdS, Strasbourg
- JL. Mandel ; IGBMC, Strasbourg
Myopathies/AFM Décrypthon:
- G. Deléage; IBCP, Lyon
- F. Desprez; ENS, Lyon
- C. Marcelle; Melbourne University, Australia
- T. Toursel; AFM, Paris
Retinal diseases:
- AMD Consortium (7 worldwide teams)
- EVI-Genoret Consortium (42 European teams)
- I. Audo; Université Pierre et Marie Curie, Paris
- S. Bhattacharya; Inst. Ophthalmology, London, UK
- J. Demongeot; U. Joseph Fourier, Grenoble
- C. Grimm; Zurich, Switzerland
- T. Léveillard; Institut de la Vision, Paris
- J. Sahel; Institut de la Vision, Paris
- C. Seitz; Institut de la Vision, Paris
- D. Zack; John Hopkins Hospital, Baltimore USA
Prostate cancer:
- J. Abecassis, IGBMC, Strasbourg
- M. Cecchini; University of Bern, Switzerland
- J. Schalken; NCMLS, Nijmegen, Netherland
- B. Wazylick; IGBMC, Strasbourg
Functional genomics data analysis:
- TF. Baumert; UdS, Strasbourg
- B. Bihain; Genclis SAS, Nancy
- A. Bloch-Zupan; IGBMC, Strasbourg
- P. Carbon, IBMC, Strasbourg
- C. da Silva; Génoscope ; Evry
- E. Friedrich; University of Luxembourg
- J. Laporte; IGBMC, Strasbourg
- A. Marchini; University of Heidelberg, Germany
- R. Romand; IGBMC, Strasbourg
- M. Roux; INIST, Nancy
- M. Sissler, IBMC, Strasbourg
- L. Tora; IGBMC, Strasbourg
- L. Vallar; CRP-Santé, Luxembourg
|
|
|
|
|
|
|